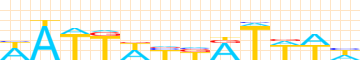
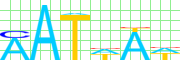
Kulakovskiy I.V., Favorov A.F., Makeev V.J. (2009) Motif discovery and motif finding from genome-mapped DNase footprint data. Bioinformatics 25(18): 2318-2325.
<up> DMMPMM motif UBX comparison [dmmpmm_compare_html]
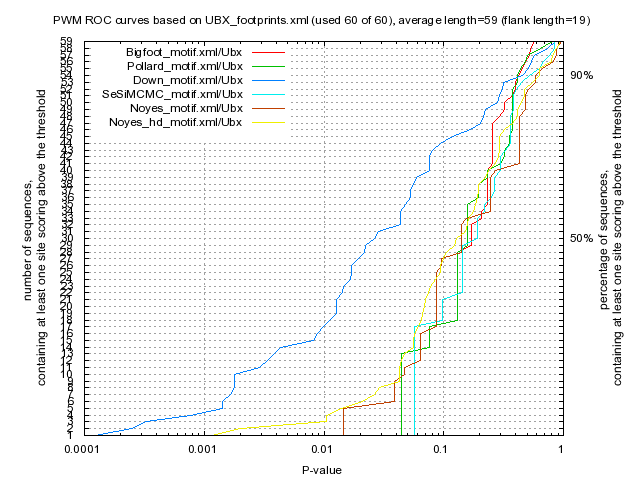 |
motif alignment
motif similarity
|
Bigfoot |
Pollard |
Down |
Bergman |
SeSiMCMC |
Noyes |
Noyes_hd |
| Bigfoot |
1.0 |
0.7637 |
0.1551 |
0.0 |
0.409 |
0.3609 |
0.2999 |
| Pollard |
0.7637 |
1.0 |
0.1616 |
0.0 |
0.4268 |
0.4355 |
0.3536 |
| Down |
0.1551 |
0.1616 |
1.0 |
0.0 |
0.2149 |
0.1744 |
0.1787 |
| Bergman |
0.0 |
0.0 |
0.0 |
0.0 |
0.0 |
0.0 |
0.0 |
| SeSiMCMC |
0.409 |
0.4268 |
0.2149 |
0.0 |
1.0 |
0.3867 |
0.3433 |
| Noyes |
0.3609 |
0.4355 |
0.1744 |
0.0 |
0.3867 |
1.0 |
0.6685 |
| Noyes_hd |
0.2999 |
0.3536 |
0.1787 |
0.0 |
0.3433 |
0.6685 |
1.0 |
|